Researchers Show How Cell-type Specific Epigenetic Clocks Could be Used to Quantify Biological Aging of Individual Cell-types
Epigenetic clocks are machine learning predictors of chronological and biological age that have found widespread application in many areas of science. They offer particular promise as a means to monitor healthy aging or to test the efficacy of anti-aging interventions in humans and animal models.
However, a major challenge in their construction and interpretation has been the underlying cell-type heterogeneity of complex tissues, since the cell-type composition of such tissues can change with age. Thus, current epigenetic clocks are composites, made up of two processes: an ‘extrinsic’ one that captures the age-associated changes in cell-type composition, and an ‘intrinsic’ one that captures the age-associated epigenetic (DNA methylation) changes that happen in individual cell-types. It is currently unknown how much of an epigenetic clock’s accuracy is due to these different extrinsic and intrinsic aging processes. Moreover, it is unclear how to quantify the intrinsic component into those of the individual cell-types, as this requires a method to quantify biological aging at cell-type resolution.
A team led by Professor Andrew Teschendorff and two of his PhD students, TONG Huige and GUO Xiaolong, from the Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences has now addressed these two outstanding questions.
Using large collections of blood and brain tissue DNA methylation datasets, the researchers demonstrated that up to 39% of an epigenetic clock’s accuracy in blood is driven by underlying changes in naïve T-cell and other immune cell fractions, whereas this extrinsic component fraction is much lower for brain tissue (12%) where it is driven by shifts in neuronal subsets.
According to Andrew Teschendorff, “We have known for a long time that aging in blood is associated with shifts in naïve T-cell fractions, but our work has now quantified how much of this extrinsic process contributes to the accuracy of epigenetic clocks”. He further pointed out that “These fractions on the extrinsic component are lower bounds, and that they are likely to rise as we have better methods to quantify cell-type fractions at higher cellular resolution”.
The researchers subsequently developed a statistical method to quantify biological aging at cell-type resolution, building cell-type specific DNA methylation clocks in two tissue-types (brain and liver), demonstrating that neuron and hepatocyte specific clocks yield more accurate predictors of chronological age in sorted neurons and hepatocytes, compared to pan-tissue clocks like the Horvath clock. In both cases, these cell-type specific epigenetic clocks also led to improved estimates of biological age compared to tissue-specific or pan-tissue clocks. For instance, the researchers found that glia specific clocks displayed a particularly pronounced age-acceleration in the temporal lobe of Alzheimer’s Disease patients, reinforcing the view that epigenetic changes in the glia compartment could be important contributors to neurodegeneration. In line with this, CpGs within these neuron and glia specific clocks displayed a significant overlap with those making up the DamAge clock, a recently published epigenetic clock whose CpGs may be causally implicated in promoting poor health outcomes.
As mentioned by the lead researcher, “It is exciting that CpGs within our neuron and glia specific clocks are mapping to genes that have been causally implicated in neurodegeneration. It shows that we are on the right track to building more meaningful molecular clocks”. However, it should be noted that many limitations remain and that further work will be necessary to explore the causal connection of age-associated epigenetic changes and neurodegeneration.
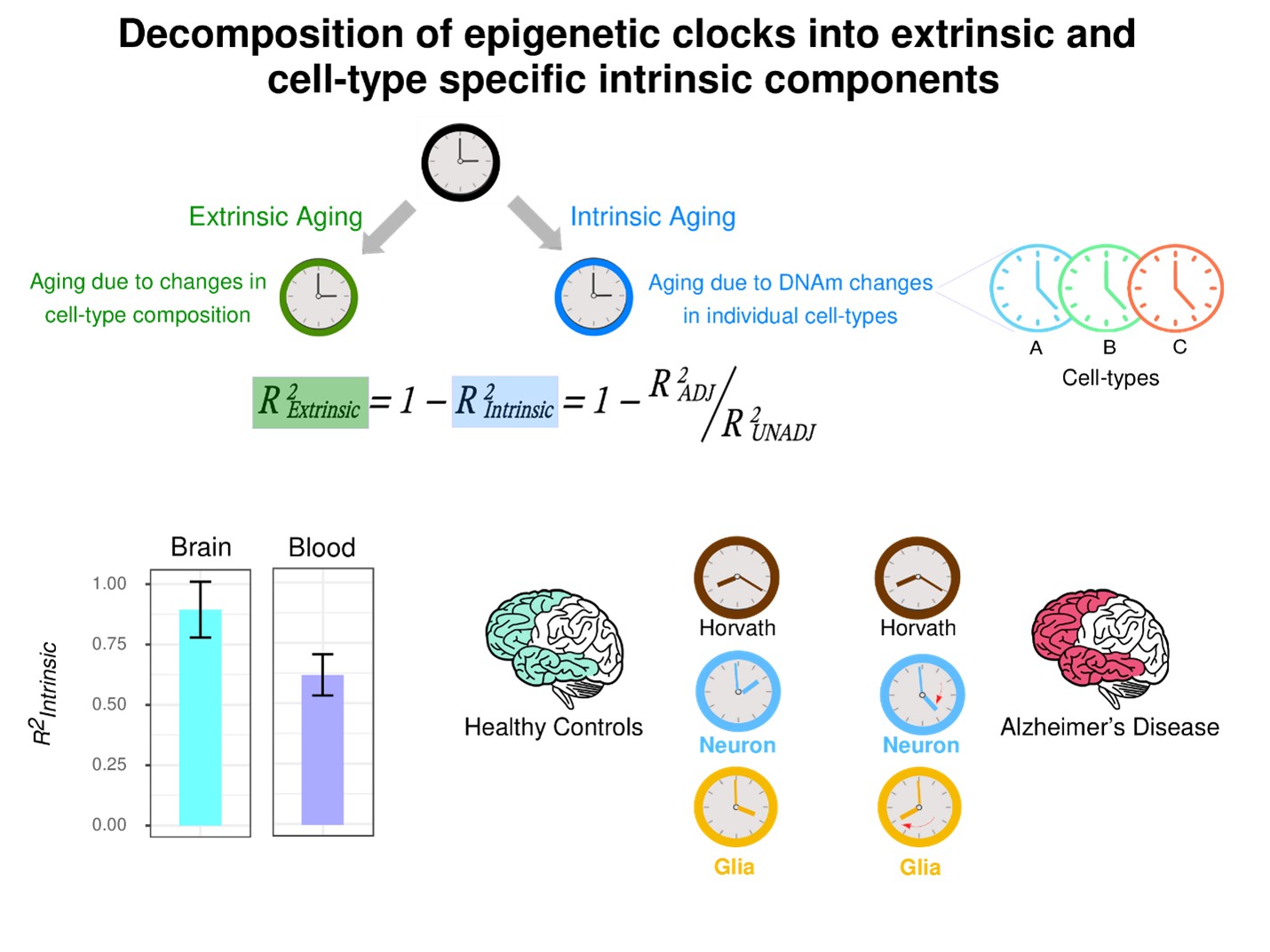
Epigenetic clocks are composites, reflecting two aging components, one reflecting age-associated changes in cell-type composition (‘extrinsic’) and another reflecting age-associated DNA methylation changes within individual cell-types (‘intrinsic’), which can also vary between cell-types, requiring the construction of cell-type specific epigenetic clocks. We quantified the intrinsic component of aging in brain and blood. In blood the extrinsic component is much larger, caused by major age-associated shifts in immune cell composition. By comparing epigenetic clock estimates in the frontal and temporal lobes of Alzheimer Disease patients to those of controls, we find that glia and neuron clocks are accelerated in AD patients, whilst the Horvath and other non cell-type specific clocks do not. The glia clock shows the strongest age-acceleration in AD. (Image by Professor Andrew Teschendorff’s group)
This work was published in the cover issue of the journal Aging, as a Priority Research Article, under the title “Cell-type specific epigenetic clocks to quantify biological age at cell-type resolution” on the 29th December 2024.
This work is funded by the Chinese Academy of Sciences and National Natural Science Foundation of China. The datasets analyzed in the manuscript are publicly available.
Scientific Contact:
Andrew E Teschendorff, Principal Investigator
CAS Key Laboratory of Computational Biology,
Shanghai Institute of Nutrition and Health,
Chinese Academy of Sciences
E-mail: andrew@sinh.ac.cn
Media Contact:
WANG Jin
Shanghai Institute of Nutrition and Health,
Chinese Academy of Sciences
Email: wangjin01@sinh.ac.cn
Web: http://english.sinh.cas.cn/
