BEACON: An Efficient and Specific Base Editor System with Low DNA Damage Response
With the collaboration of a group of Chinese scientists, a new type of dCas12a-derived bases editors, named BEACONs, has been recently developed to achieve efficient and specific editing with only low levels of DNA damage response. The corresponding paper entitled “Cas12a Base Editors Induce Efficient and Specific Editing with Low DNA Damage Response” was published online in Cell Reports on June 2, 2020. This research was led by Dr. YANG Li at CAS-MPG Partner Institute for Computational Biology, Shanghai Institute of Nutrition and Health of Chinese Academy of Sciences, Dr. CHEN Jia at the School of Life Science and Technology, ShanghaiTech University, and Dr. LIU Zhen at Institute of Neuroscience of Chinese Academy of Sciences.
The CRISPR/Cas9 system has been successfully applied in various living organisms for genome editing. The Cas9 nuclease generates DNA double-strand breaks (DSBs) at specific genomic loci under the direction of guide RNAs (gRNAs), which are further processed by downstream DNA repair pathways to induce gene editing outcomes. Therefore, the formation of DSBs is required for CRISPR-Cas9-mediated genome editing. Although Cas9 nickase (nCas9) only generates DNA single-strand breaks(SSBs), apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC)/activation-induced cytidine deaminase(AID) family members together with base excision repair proteins can lead to DSBs during the repair of these SSBs. Similarly, base editors (BEs) that link nCas9 with APOBEC1 are also associated with DSBs, demonstrated by the induced insertions or deletions (indels) around BE-target sites.
In this study, researchers show that the original version of catalytically dead Cas12a (dCas12a)-conjugated BEs induce a basal level of DNA breaks and minimally activate DDR proteins, including H2AX, ATM, ATR, and p53. By fusing dCas12a with engineered human apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A (APOBEC3A), researchers further develop the BEACON (Base Editing induced by human APOBEC3A and Cas12a without DNA break) system to achieve enhanced deamination efficiency and editing specificity. Efficient C-to-T editing is achieved by BEACON in mammalian cells at a level comparable to AncBE4max, with only low levels of DDR and minimal RNA off-target mutations. Importantly, BEACON induces in vivo base editing in mouse embryos, and targeted C-to-T conversions are detected in F0 mice.
During this study, to precisely and efficiently evaluate RNA OT mutations in the application of BEs, researchers also developed an powerful RNA-editing analysis-pipeline to decode all-twelve-types of RNA-editing (RADAR, https://github.com/YangLab/RADAR). By using RADAR, researchers confirmed that BEACON only induced a background level of RNA C-to-U OT mutations.
This study was supported by grants from the National Natural Science Foundation of China, Ministry of Science and Technology, Shanghai Municipal Science and Technology Commission, etc.
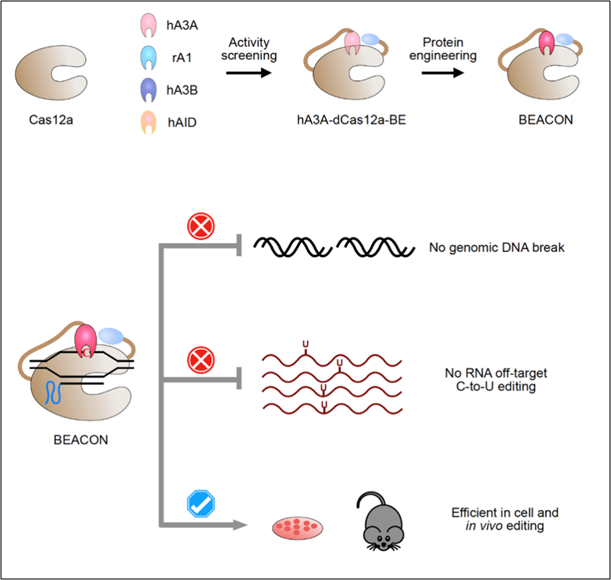
The development and applications of BEACON. (Image provided by Dr. YANG Li's Group)
Media Contact:
WANG Jin (Ms.)
Shanghai Institute of Nutrition and Health,
Chinese Academy of Sciences
Email: sibssc@sibs.ac.cn
