- Home >> ALL News >> Highlights
Researchers Reveal Novel Mechanism Behind Host and Microbiota Interaction
A research team led by Prof. QIAN Youcun from the Shanghai Institute of Nutrition and Health (SINH), Chinese Academy of Sciences (CAS), and Prof. SONG Xinyang from the Center for Excellence in Molecular Cell Science, CAS, has uncovered a novel mechanism by which apolipoprotein-dependent recognition of sphingolipids mediates host–microbiota interactions and regulates intestinal homeostasis. Their groundbreaking findings entitled "Targeting symbionts by apolipoprotein L proteins modulates gut immunity" were published in Nature on May 14, 2025.
The mammalian gut is a key site of host–microbiota interaction, where commensal microbes influence immune cell function through metabolites and structural molecules. These signals can promote Treg differentiation or IEL accumulation. The host maintains microbiota balance via broad defense mechanisms such as mucus, AMPs, sIgA, and complement. However, whether the host can selectively recognize and regulate specific microbes remains unclear. Uncovering this could reveal how precise immune mechanisms shape gut symbiosis and inform targeted therapies for microbiota-associated diseases.
To systematically identify key host-derived molecules involved in regulating gut microbiota, the researchers first performed high-throughput proteomic analysis to compare the protein composition of the ileal mucus layer between conventionally raised (CR) and germ-free (GF) mice. By screening proteins that were significantly upregulated in the presence of microbiota and combining this with a proteomic analysis of host proteins bound to commensal bacteria isolated from the ileum of CR mice, the team identified a previously uncharacterized member of the apolipoprotein family—Apolipoprotein L9a/b (APOL9a/b, or APOL9). They found that APOL9 expression was notably elevated in CR mice and was primarily derived from intestinal epithelial cells (IECs).
To explore whether APOL9 targets specific microbial groups, the researchers developed an "APOL9-seq" strategy by combining flow cytometric sorting with 16S rRNA gene sequencing. This analysis revealed that both murine APOL9a/b and its human homolog APOL2 displayed striking specificity, almost exclusively binding to bacteria of the order Bacteroidales, in both mouse and human systems. To uncover the molecular basis of this specificity, the researchers used Bacteroides thetaiotaomicron, a representative commensal bacterium from the Bacteroides genus, as a model strain. Using genetic editing tools, they knocked out genes involved in membrane lipid metabolism and found that APOL9/2 binding required a specific lipid on the bacterial membrane—ceramide-1-phosphate (Cer1P). Deletion of the key enzyme involved in Cer1P biosynthesis significantly impaired the binding of APOL9/2 to the bacteria, identifying Cer1P as the critical molecular signature recognized by the host. This finding demonstrates, for the first time, that the host can selectively label specific microbial taxa by recognizing unique lipid structures of commensal bacteria.
Unlike conventional antimicrobial peptides that kill target microbes, APOL9 binding does not damage or lyse the bacteria. Instead, it non-lethally induces the release of a large quantity of outer membrane vesicles (OMVs) from the targeted Gram-negative bacteria. These nanosized OMVs, ranging from tens to hundreds of nanometers in diameter, are enriched with bacterial-derived lipids, proteins, and polysaccharides, making them recognizable by the host immune system. Functional analyses revealed that these APOL9-induced OMVs are not only captured by the host but also actively enhance local immune responses. Specifically, they activate interferon-gamma (IFN-γ) signaling, significantly upregulating the expression of major histocompatibility complex class II (MHC-II) molecules in IECs, thereby promoting the development and maintenance of a specialized population of immunoregulatory CD4⁺CD8αα⁺ T lymphocytes within the intestinal epithelium.
To functionally validate the role of APOL9, the researchers generated Apol9a/b double-knockout mice and subjected them to oral infection with Salmonella typhimurium. These knockout mice exhibited significantly higher pathogen loads in the gut, increased dissemination of bacteria to visceral organs such as the liver and spleen, and a higher risk of systemic infection and mortality. In contrast, supplementation with OMVs derived from APOL9-stimulated bacteria significantly alleviated infection symptoms and improved host immune defense.
This study is the first to reveal how a host protein can specifically recognize commensal microbial membrane lipids and induce the release of beneficial immunomodulatory OMVs. It provides a new theoretical framework for understanding how the host actively shapes the gut microbiota and lays a molecular foundation for developing next-generation microbiota-targeted immune interventions.
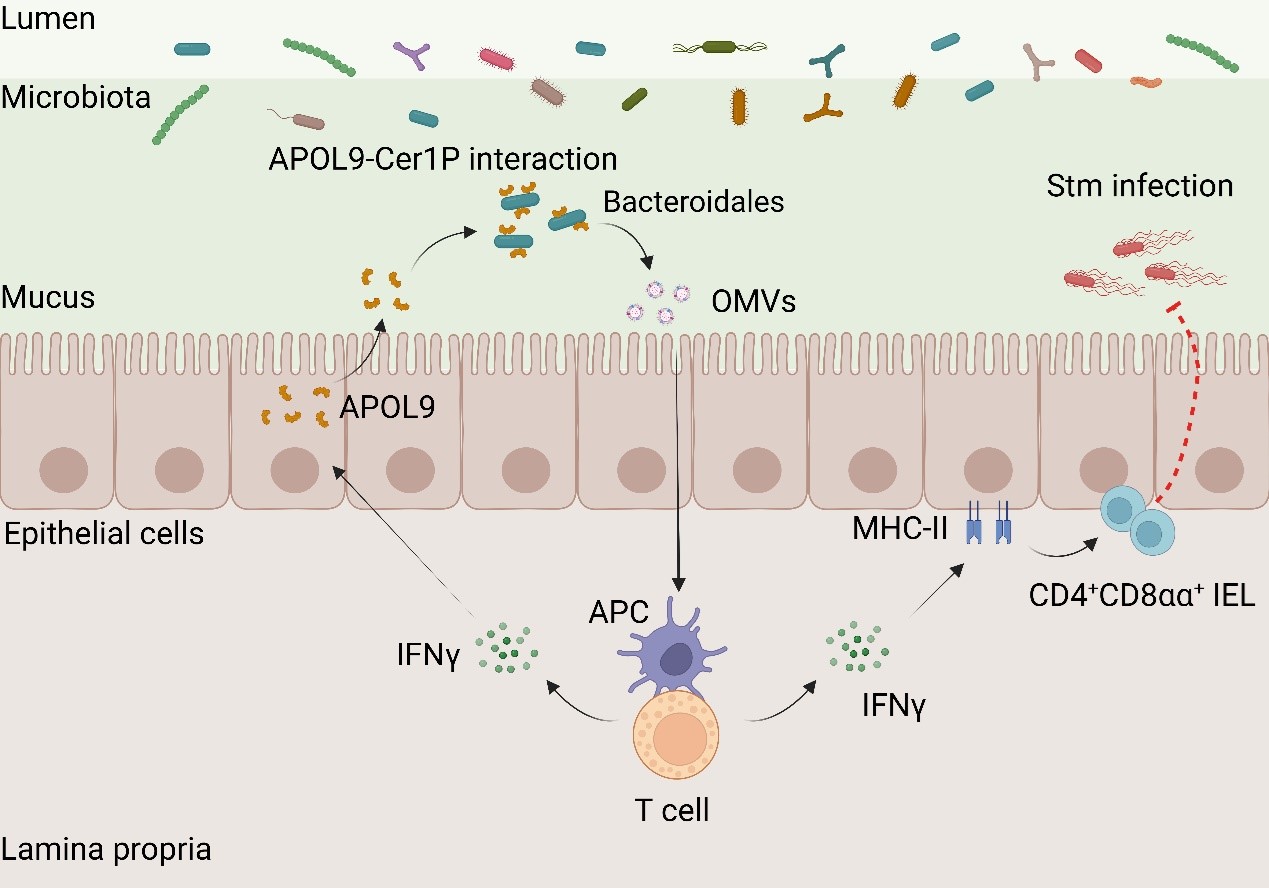
The proposed model for the novel apolipoprotein APOL9 that enhances mucosal immunity by activating the IFN-γ-MHC-II pathway through induction of OMVs release in Bacteroidales.
(Image provided by Prof. QIAN Youcun's group)
This work was also supported and assisted by Prof. Dennis L. Kasper from Harvard Medical School, Prof. LIANG Guanxiang from Tsinghua University School of Medicine, Prof. SHI Jiantao from the CAS Center for Excellence in Molecular Cell Science and Prof. HU Guohong from SINH. The study was funded by the National Key R&D Program of the Ministry of Science and Technology, the National Natural Science Foundation of China, and supported by the institutional core facilities and animal platform of SINH.
Paper link: https://doi.org/10.1038/s41586-025-08990-4
Scientific Contact:
Prof. QIAN Youcun
Shanghai Institute of Nutrition and Health,
Chinese Academy of Sciences
Email: ycqian@sinh.ac.cn
Media Contact:
WANG Jin
Shanghai Institute of Nutrition and Health,
Chinese Academy of Sciences
Email: wangjin01@sinh.ac.cn
Web: http://english.sinh.cas.cn/