- Home >> ALL News >> Highlights
DNA Base Editing Induces Substantial Off-target RNA Mutations and Their Elimination by Mutagenesis
A recent study published in Nature demonstrated that DNA base editors generated tens of thousands of off-target RNA single nucleotide variants (SNVs) and these off-target SNVs could be eliminated by introducing point mutations to the deaminases. This work revealed a previously overlooked aspect of the risk of DNA base editors, and also provided a solution to the problem by engineering deaminases.
This work was performed by researchers from Dr. YANG Hui’ s Lab at the Institute of Neuroscience, Chinese Academy of Sciences (CAS), and Dr. LI Yixue’s team at CAS-MPG Partner Institute for Computational Biology, Shanghai Institute of Nutrition and Health of CAS and Dr. GUO Fan’s team at Sichuan University.
DNA base editing methods have enabled direct point mutation correction in genomic DNA without generating any double-strand breaks (DSBs), but the potential off-target effects have limited the application of these methods. Adeno-associated viruses (AAV) are the most common delivery system for DNA editing gene therapies, and these viruses can sustain long-term gene expression in vivo, so the extent of potential RNA off-target effects induced by DNA base editors is of great concern for their clinical application.
Several previous studies have evaluated off-target mutations in genomic DNA, while the deaminases integral to commonly used DNA base editors often exhibit RNA binding activities. For example, the cytosine deaminase APOBEC1 used in cytosine base editors (CBEs) was found to target both DNA and RNA, and the adenine deaminase TadA used in adenine base editors (ABEs) was found to induce site-specific inosine formation on RNA. However, any potential RNA mutations caused by DNA base editors has not been evaluated.
In order to evaluate the off-target effect of DNA base editors at the level of RNAs, the researchers quantitively counted the off-target RNA SNVs in each replicate of CBE or ABE treated cells. They next explored the possibility of elimination of the off-target RNA SNVs by engineering deaminases of DNA base editors.
The researchers transfected one type of CBEs, BE3 (APOBEC1-nCas9-UGI) or ABEs, ABE7.10 (TadA-TadA*-nCas9), together with GFP and with or without single guide RNA (sgRNA), into HEK293T cultured cells. After validating the high on-target efficiency of DNA editing by both BE3 and ABE7.10 in these HEK293T cells, they performed RNA-seq at an average depth of 125X on these samples and quantitively evaluated the RNA SNVs in each replicate.
The on-target editing efficiency was evaluated in each replicate of the CBE or ABE treated cells to guarantee the efficient editing. Then the number of off-target RNA SNVs in CBE and ABE treated groups were compared with the GFP-only control group and they found strikingly higher numbers of RNA SNVs in DNA base editor treated cells. Besides, they also found the mutation bias in BE3 or ABE7.10 treated cells were same as that of APOBEC1 or TadA, respectively, indicating the off-target effects were caused by the overexpression of DNA base editors. In addition, they found CBE and ABE specific motifs and genetic regions of these off-target RNA SNVs.
To eliminate the RNA off-target activity of base editors, the researchers examined the effect of introducing point mutations on APOBEC1 or TadA. Three high fidelity variants BE3W90Y+R126E, BE3 (hA3AR128A) and BE3 (hA3AY130F) reduced RNA off-target SNVs to a base level. Similarly, an ABE variant ABE7.10F148A also showed complete elimination of off-target effects.
In this project, the researchers revealed a risk of DNA base editors which was previously overlooked. By introducing point mutations to the deaminases, high-fidelity variants for both CBEs and ABEs were obtained. This research also provided a suggestive method by rational engineering to increase the specificity of base editors.
This work entitled “Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis” (https://doi.org/10.1038/s41586-019-1314-0) was published online in Nature on June 10, 2019. ZHOU Changyang, SUN Yidi, YAN Rui, LIU Yajing and ZUO Erwei are the first authors with equal contribution. This work was supported by grants of R&D Program of China, CAS Strategic Priority Research Program, National Natural Science Foundation of China, Shanghai Municipal Science and Technology Major Project, Shanghai City Committee of Science and Technology Project, National Science and Technology Major Project, and National Key Research and Development Program of China.
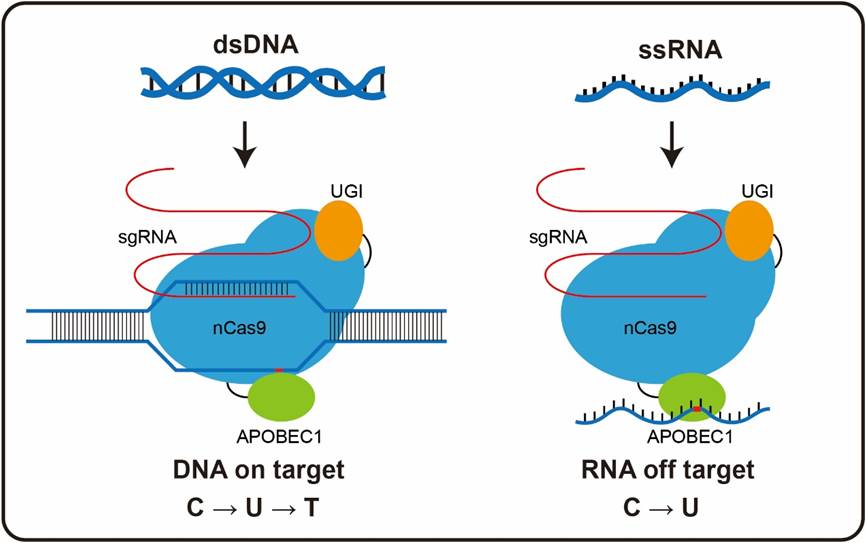
This study obtained both high-fidelity variants for both CBEs and ABEs by introducing point mutations to the deaminases and provided a suggestive method using rational engineering to increase the specificity of base editors.(Image provided by Dr. LI yixue's group)
Media Contact:
WANG Jin (Ms.)
Shanghai Institute of Nutrition and Health,
Chinese Academy of Sciences
Email: sibssc@sibs.ac.cn