Comprehensive Base Editing Mediated by hAPOBEC3A-conjugated Base Editors
Base editors (BEs) are developed by combining different nucleotide deaminase family members, including cytidine deaminase family members (e.g., APOBECs) and adenosine deaminase family members (e.g., Adenosine deaminases acting on RNA, ADARs), with the CRISPR-Cas system (e.g., CRISPR/Cas9 and CRISPR/Cpf1). Various BEs have been used for targeted C-to-T/A-to-G base editing in different species. Numerous human diseases have been reported to be driven by point mutations in genomic DNA. With recently developed BEs, these disease-related point mutations can be potentially corrected, providing new therapeutic options. The newly-developed base editing technology has been highlighted by Science as one of its top 10 “breakthrough in 2017”.
A team of scientists led by Dr. CHEN Jia, Dr. HUANG Xingxu of the School of Life Science and Technology at ShanghaiTech University and Dr. YANG Li of the CAS-MPG Partner Institute for Computational Biology at the Chinese Academy of Sciences, have developed a series of novel base editors based on a spectrum of cytidine deaminases from different species, including human apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC3A, hA3A) cytidine deaminase family members.
By analyzing disease-related T-to-C mutations that can be theoretically reverted to thymines by BEs, the research team found that ~ 43% of them are on cytosines in the context of CpG dinucleotides. It is well known that C of CpG is usually methylated in mammalian cells, and methylation of C strongly suppresses cytidine deamination activity of some APOBEC/AID deaminases. Consistently, previously developed BEs that are based on rat APOBEC1 (rA1) cytidine deaminase are inefficient in editing cytosines in highly-methylated regions. To develop BEs for efficient C-to-T base editing in highly methylated regions, researchers developed a series of BEs by fusing Cas9 nickase with a dozen of different APOBEC/AID deaminases, and then showed that most of them can be used for C-to-T base editing.
Importantly, a novel BE based on hA3A (hA3A-BE) and its engineered versions with narrower editing windows can mediate efficient C-to-T base editing in regions with high methylation levels and other examined regions with different contexts. Overall, these newly developed hA3A-BEs can comprehensively induce efficient base editing in all examined contexts, including both methylated DNA regions and GpC dinucleotides.
The research findings entitled “Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion” was published online in Nature Biotechnology on August 20, 2018.This study was supported by grants from the National Natural Science Foundation of China, Ministry of Science and Technology, Shanghai Municipal Science and Technology Commission, and ShanghaiTech University.
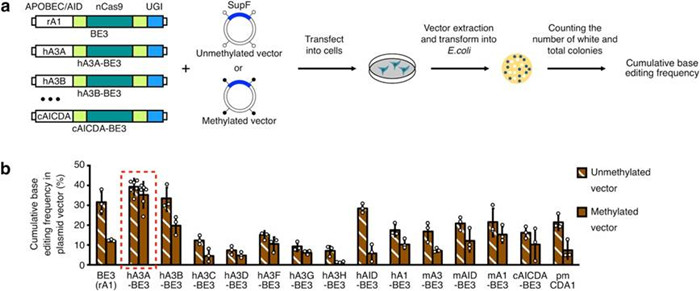
Screening for high-efficient base editors on methylated regions.
(a) Construction of various BEs and screening for efficient base editing in highly methylated regions.
(b) hA3A-BE3 can induce efficient base editing in highly methylated regions.
(Image by Dr. CHEN Jia, Dr. HUANG Xingxu and Dr. YANG Li’s teams)
WANG Jin (Ms.)
Shanghai Institute of Nutrition and Health,
Chinese Academy of Sciences
Email:
