Scientists Release a Database for Genomic Diversity and Genetic Ancestry of Global Human Populations
Population genetic diversity is shaped by complex process across human evolutionary history, including population divergence, migration, isolation, and admixture. Local adaptations to environments such as high-altitude, pathogen and dietary shift, could also affect the diversity of human genome locally. Genomic diversity further determines largely the phenotypic diversity. Therefore,understanding phenotypic differences between populations (including diseases, anthropological traits and adaptive traits) relies on delineating genetic diversity and ancestry as well as evolutionary history of human populations.
A new study led by Prof. Dr. XU Shuhua entitled “PGG.Population: a database for understanding the genomic diversity and genetic ancestry of human populations” was published online in Nucleic Acids Research on Nov. 3rd, 2017. It analyzed the genomic diversity and genetic ancestry of 356 ethnic groups from 107 counties, and released a database named “PGG.Population” (www.pggpopulation.org) through which users can freely access to the data and related information of global populations. PGG.Population is the only databasebeing committed to dissect the genetic affinities of populations and genetic ancestries for each ethnic group at the genomic level; it is also the one covering the largest number of populations to date, proving a query and analysis platform for researchers, clinical and medical doctors, students and the public to understand the genetic background of each ethnic group.
Over the past decades, many joint forces based on international collaborations have made remarkable achievements in studying human genetic variation, such as the Human Genome Diversity Project, the HapMap Project, the HUGO Pan-Asian SNP Project and the 1000 Genomes Project. Nonetheless, most efforts have focused on major groups of large population size but ignored indigenous groups which usually have much smaller population size. PGG.Population was born at the right moment. Researchers sequenced and collected the genomes of diverse populations especially for small/unique groups, integrated them and re-analyzed each combined dataset. Meanwhile, they built a free available database to illustrate the genomic diversity and genetic ancestry (including basic information, Y and mtDNA haplogroup, genetic affinities, population structure, admixture and local adaptation) for each ethnic group. Up to date, the databasehas documented 7122 genomes, representing 356 non-redundant populations/groups from 107 countries in 8 regions (Africa, America, Central Asia and Siberia, East Asia, Oceania, South Asia, Southeast Asia, and West Eurasia) collected from 27 studies. Every population has its own story. The long-term ambition of the PGG.Population, together with the joint efforts from other researchers who contributed their data to our database, is to create a comprehensive depository of geographic and ethnic variation of human genome, as well as a platform bringing influence on future practitioners of medicine and clinical investigators.
However, it still has a long way to achieve the goal. There are more than 2,000 populations worldwide, and the database currently only covers one sixth of those groups. Scientists are now sequencing and collecting genomes from more populations. In addition, PGG.Population will extend to comprehensive analysis of population genetics and genomics. “We call for more cooperation and contribution from geneticists, linguists, anthropologists, medical and clinical doctors”, said Dr. XU Shuhua, Principal Investigator and Group Leader of Max Planck Independent Research Group on Population Genomics, CAS-MPG Partner Institute for Computational Biology (PICB), Shanghai Institutes for Biological Sciences (SIBS), Chinese Academy of Sciences (CAS).
This work was supported by the Strategic Priority Research Program and Key Research Program of Frontier Sciences of the Chinese Academy of Sciences (CAS), the National Natural Science Foundation of China (NSFC) grant, the National Science Fund for Distinguished Young Scholars, the Program of Shanghai Academic Research Leader, and the National Key Research and Development Program, etc.
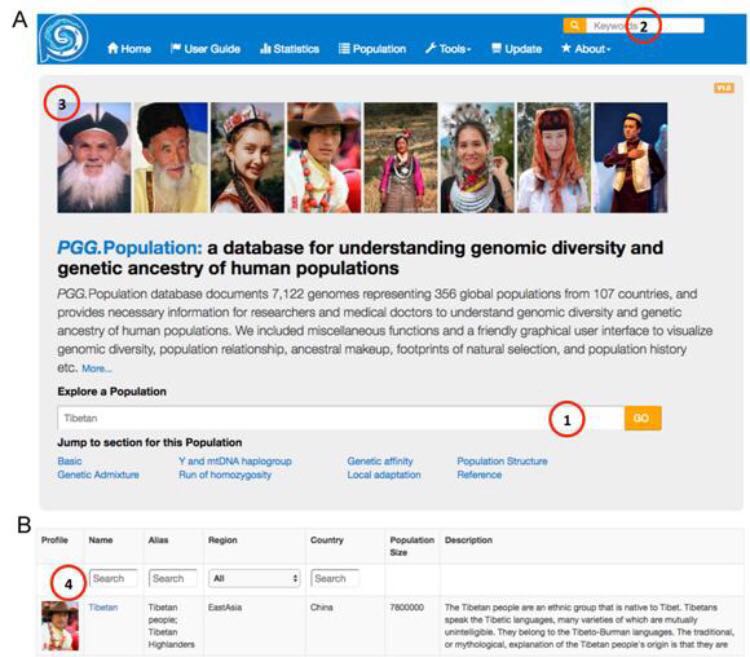
Interface of “PGG.Population” Database
(Image provided by Dr. XU Shuhua's Group)
CONTACT:
Prof. Dr. XU Shuhua, Group Leader & Principal Investigator
Max Planck Independent Research Group on Population Genomics,
CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
Email: xushua@picb.ac.cn
Tel: +86-21-54920479
